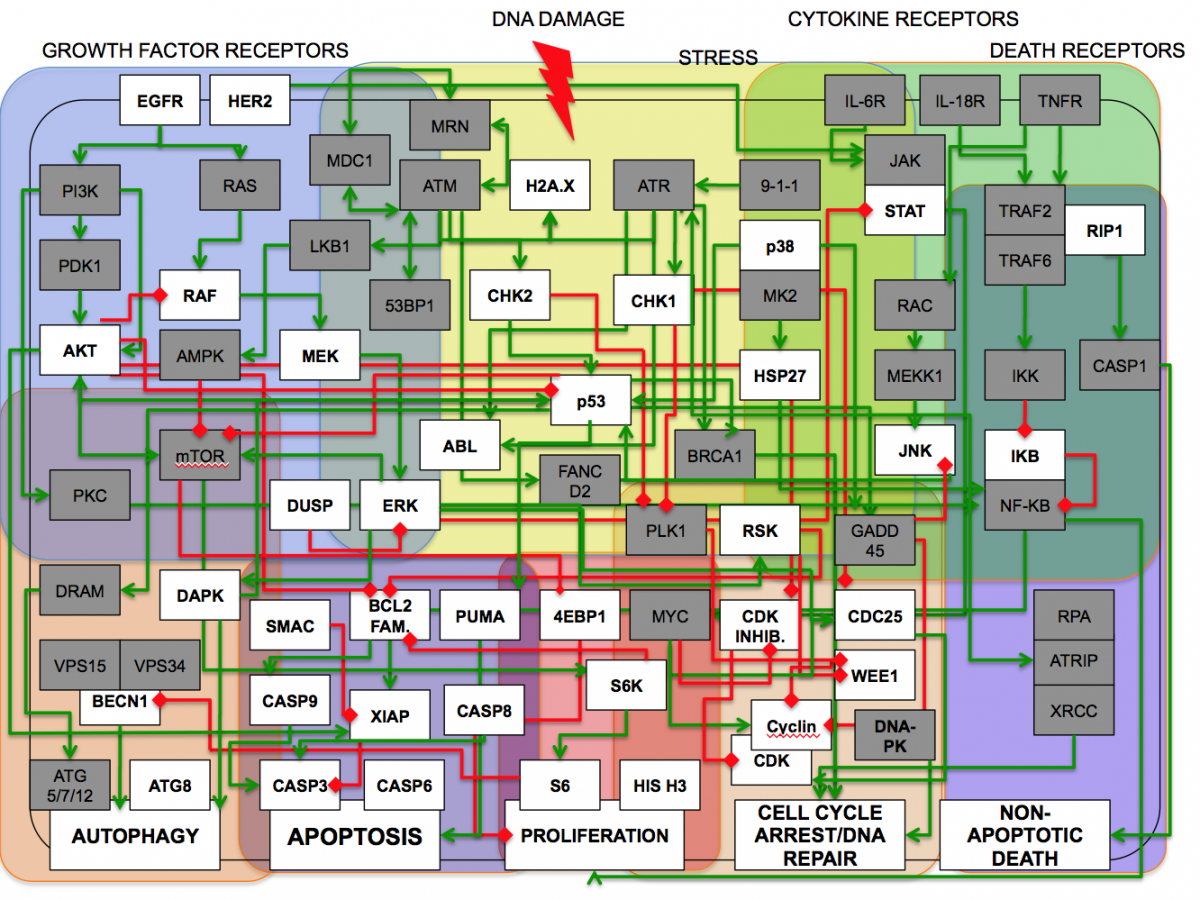
When cells encounter stress or injury such as DNA damage or infection, they activate complex signaling networks that regulate their ability to repair the damage and return to a homeostatic equilibrium. These networks must integrate a wide variety of signals to ultimately control cell cycle arrest or progression, coordinately regulate specific patterns of gene expression and/or initiate programmed cell death. Protein kinases play a pivotal role in these signaling pathways, and alterations in their normal function contributes to tumor development and progression. Intentional targeting of these pathways can also enhance chemotherapeutic drugs and radiation to kill cancer cells.
DNA damage signaling pathways are intimately interconnected with pathways involved in cell growth, RNA processing and cytokine signaling, but how these pathways are integrated together at the systems level is poorly understood. Cells and tissues subjected to other types of injuries and/or infection by pathogens activate many of the same pathways involved in the DNA damage response. Inappropriate activity of these pathways cause auto-inflammatory tissue damage and multiple organ failure in states of overwhelming infection and sepsis as a result of mis-regulation of cytokine feedback loops, dysfunction of the blood clotting cascade, and uncontrolled activity of neutrophils, macrophages and lymphocytes.
How are the signals from individual pathways integrated at the molecular level to control the phenotypic response of cells to infection, stress and DNA damage? What are the key pathways and molecules that are involved in these cellular events, and how are their activities and their interactions regulated by protein phosphorylation? How can these pathways be therapeutically manipulated using combination chemotherapies to re-wire tumor cells for optimal killing, or to limit cytokine-mediated inflammation and death? Our lab uses a broad range of technologies to decode how these cell signaling pathways are “wired” into functional networks through proteomic methods, high- and medium-throughput signaling assays, RNAi-based screens using high-content imaging, and computational/bioinformatics approaches, together with more traditional techniques from cell biology, physical biochemistry, structural biology and mouse genetics. We also have a long history of inventing new chemical, biochemical, and computational methods to study signaling, including peptide library-based screens and motif-based bioinformatics tools for building signaling networks in silico.
If you have interest in becoming a postdoctoral fellow in the Yaffe lab, send your C.V. and cover letter expressing your interests as they reflect on the direction of the laboratory.
Media
Solutions with/in/sight: Masterclass with Michael Yaffe, November 2, 2016

